Pharmacophore generation
This protocol generate a Pharmacophore object that can be parse by RDKit from a
SetOfSmallMolecules. If the input molecules are docked to a structure, the output pharmacophore keeps that structure
too. The different features that can be captured by the pharmacophore are defined in the BaseFeatures.def file of RDKit.
scipion-chem uses the families of those features as own features (Donor, Acceptor, Hydrophobe, Aromatic…).
Different cluster parameters can be chosen to generate the pharmacophore, they control the clustering method and how conserved the feature must be among the docked small molecules.
Input
Note
All parameters include a help button that gives further information for each of them.
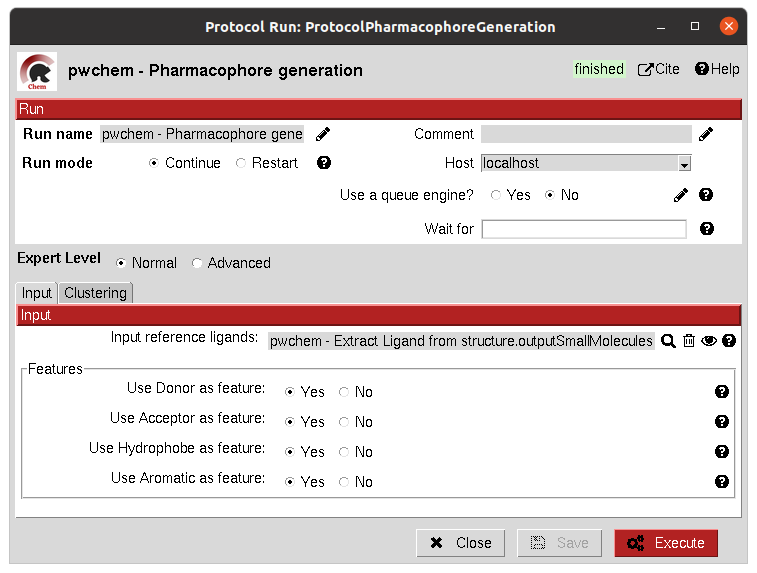
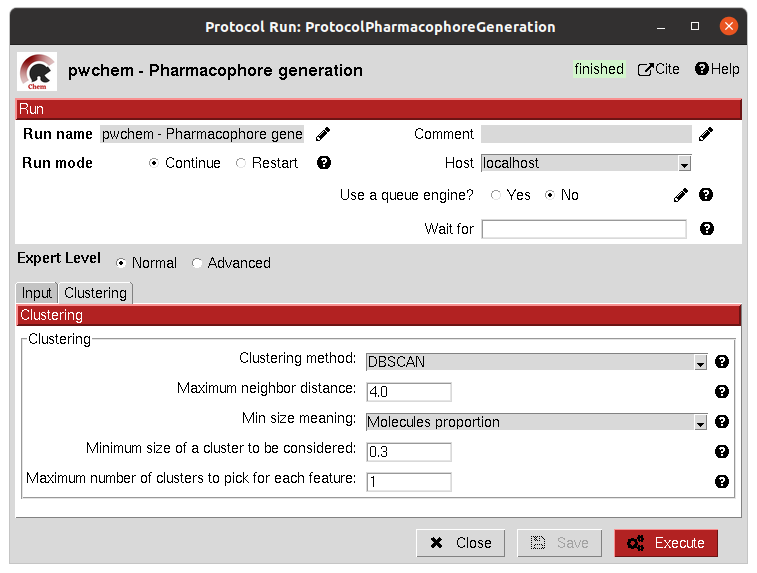
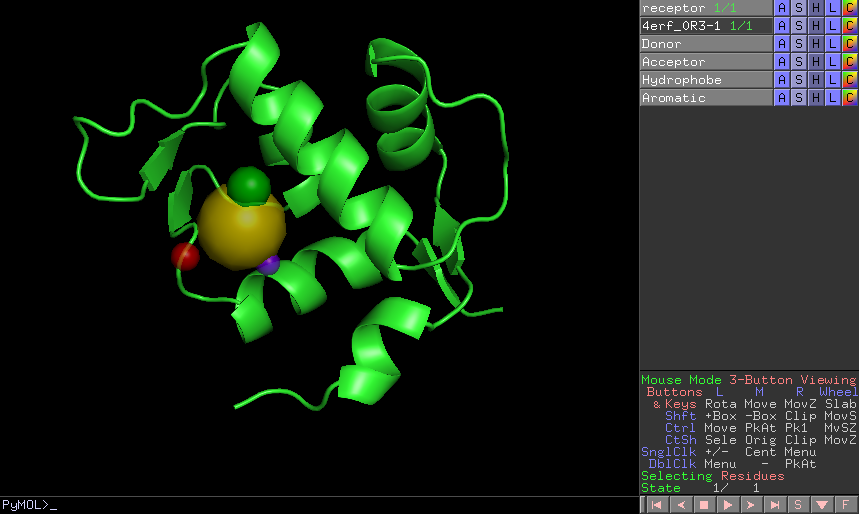
The result of this protocol is a PharmacophoreChem object containing the extracted features from the set of small molecules
with the specified clustering parameters.
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests pwchem.tests.tests_pharmacophores.TestPharmGeneration