Consensus docking
This protocol performs a consensus operation over several docked SetOfSmallMolecules, studying which positions are
shared among all or a subset of the input sets. Similarly to the Consensus structural ROIs
protocol, it might be used to obtain the most robust results, this time out of different docking protocols.
The clustering of the positions is performed based on their RMSD and different options can be chosen. The default option will use scipy package for the clustering, allowing parallelization and using an optimized code. However, due to the quadratic nature of the problem, this might be too computationally expensive, so we offer another option where the clusters are formed calculating only the distance to each cluster representative. This representative is the molecule of the cluster that has the smallest energy or biggest score. The clustering using this method is not as robust and will depend on the molecules order, but the problem will no longer be quadratic. Choose your best option wisely.
Input
Note
All parameters include a help button that gives further information for each of them.
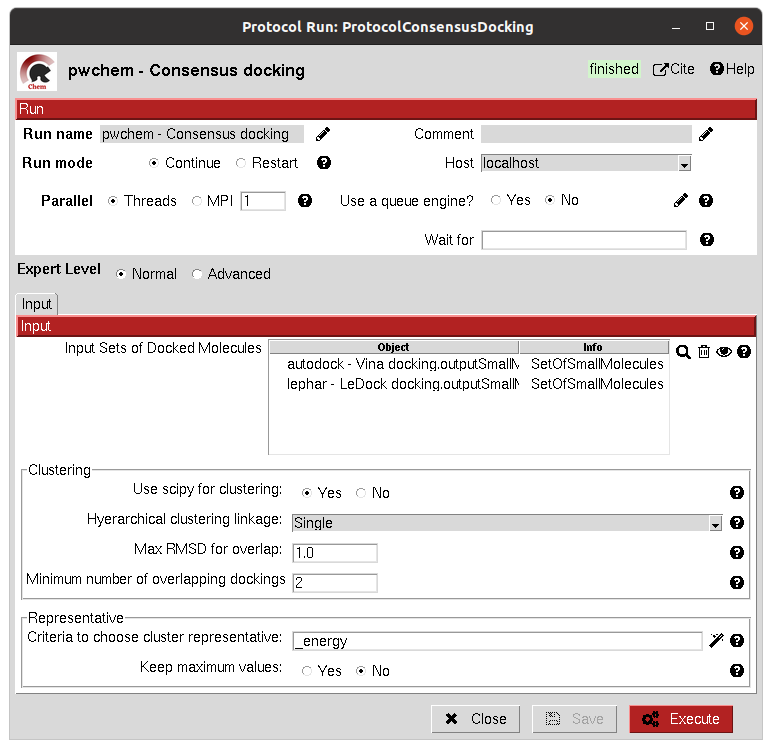
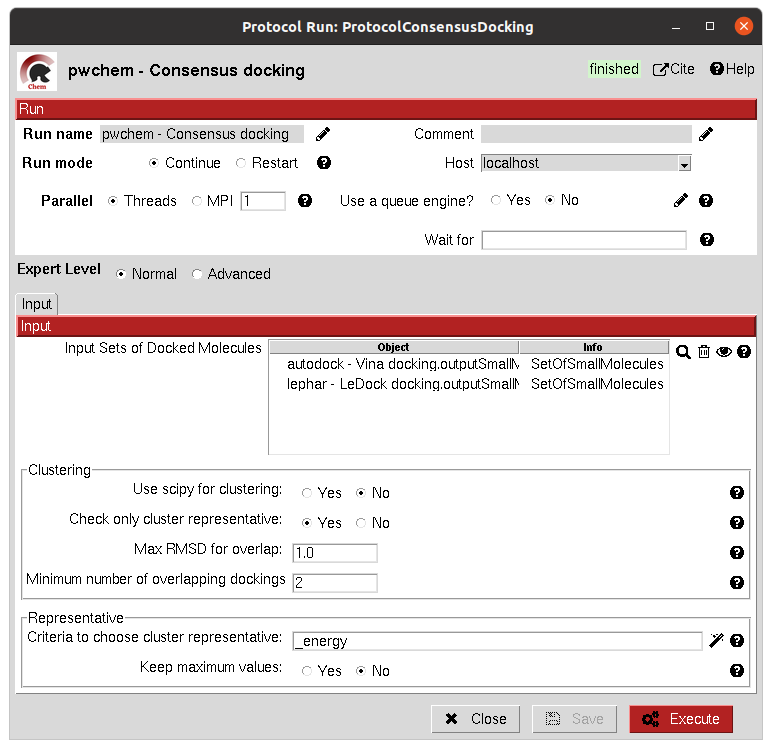
The result of this protocol is a SetOfSmallMolecules with the consensus docking positions.
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests pwchem.tests.tests_docking.TestConsensusDocking