Pairwise Alignment
This protocol performs a pairwise alignment using clustal omega over two input sequences.
These sequences can be input either from a Sequence or an AtomStruct object, in the later,
the chain must also be specified.
Input
Note
All parameters include a help button that gives further information for each of them.
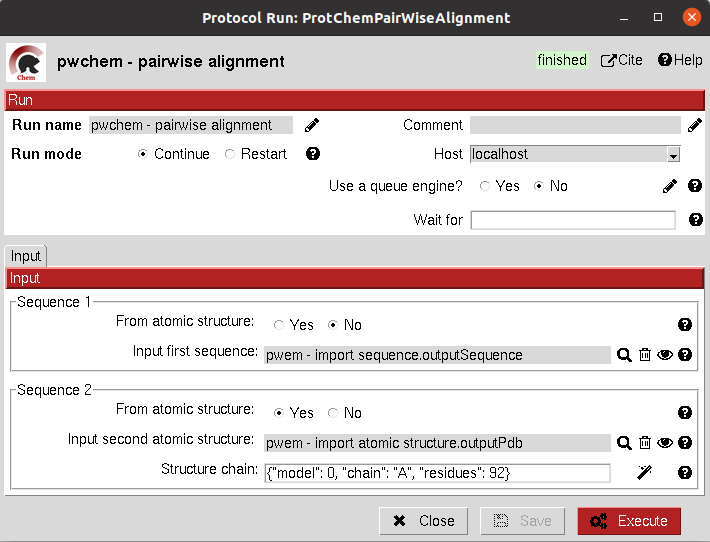
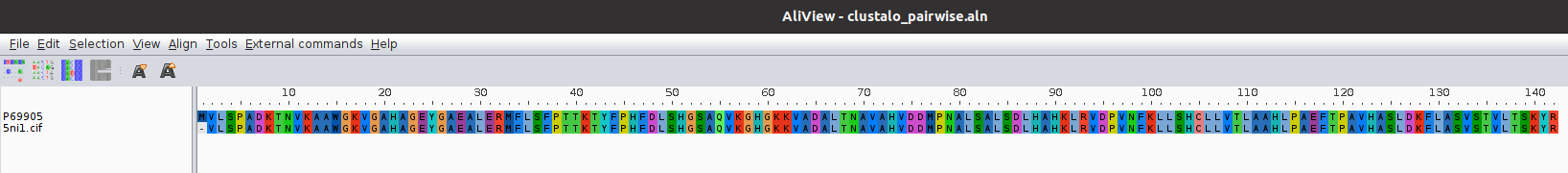
The result of this protocol is a SetOfSequences with the two input sequences aligned.
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests pwchem.tests.tests_sequences.TestPairwiseAlign