Mutation modelling
This protocol follows the Modeller wiki for single residue mutation modelling and energy optimization. The protocol includes different sections where you can define the parameters you want to use for the modelling:
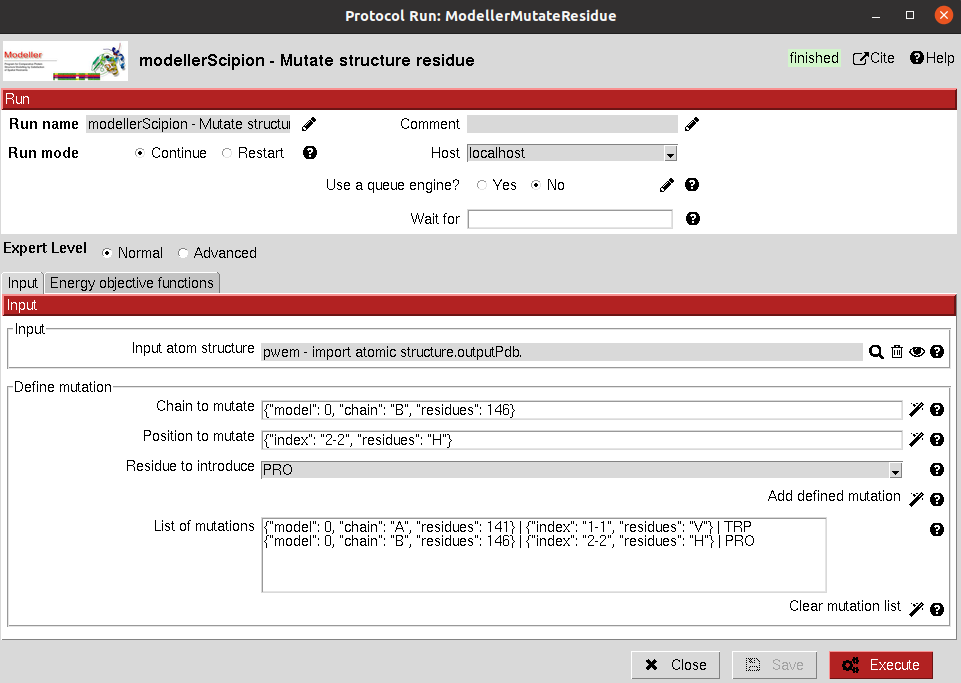
Input
Input: It must be an
AtomStructobject containing the protein structure you want to mutate.Define mutation: You can define the residue in the structure you want to mutate. Using the wizards, you can choose the chain and position from the structure. The third wizard will save the mutation in the list and they will be performed sequentially.
Distance parameters: The different distances using for determining the interactions can be tuned.
Energy restraints: Define different restraints on the energy calculations.
Note
All parameters include a help button that gives further information for each of them.
The result of this protocol is an AtomStruct object with the mutated model.
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests modellerScipion.tests.test_mutate_residue.TestModellerMutateRes