LeDock Docking
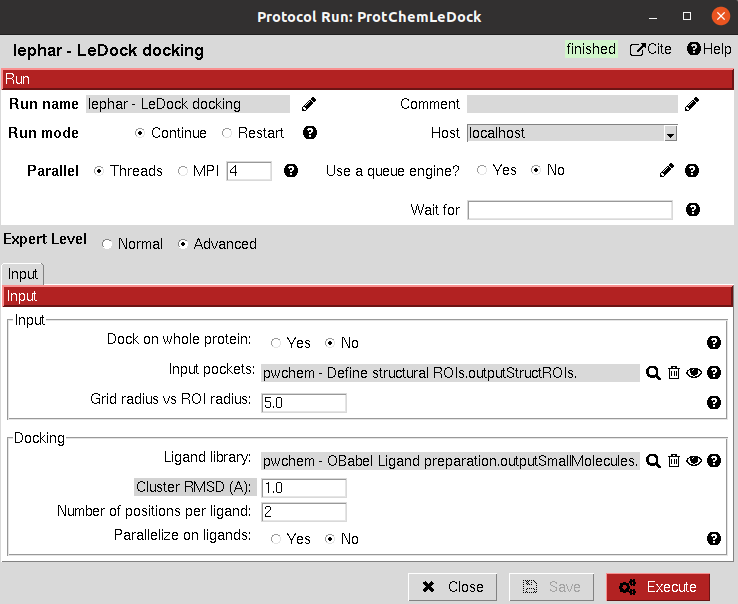
This protocol performs docking using LeDock. As for the rest of docking programs in Scipion Chem, the docking can be
performed either in the whole structure or on a SetOfStructROIs. Then, the second part of the input is the
SetOfSmallMolecules which will be docked. The docking can be paralelized over the ligands (by default, it is also done
over the StructROIs). Also, an RMSD threshold can be set to automatically discard very similar poses.
Input
Note
All parameters include a help button that gives further information for each of them.
The result of this protocol is a SetOfSmallMolecules containing the docked ligands onto the protein structure.
These molecules can be visualized clicking on Analyze Results, which will open a for to select visualization of the
whole set, individual StructROIs, all poses of a molecule, single molecules or single molecules using PLIP
(checking interactions).
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests lephar.tests.test_ledock.TestLeDock