BLAST search
This protocol performs a BLAST search over a database, which can be local or a web-server BLAST database.
Input
Note
All parameters include a help button that gives further information for each of them.
The input of the protocol is a sequence, which can be from a protein or nucleotide, and the user will be able to define the type of search (blastp, blastn, blastx, …).
In the second parameters tab, different parameters for the search can be tuned. If you are not sure of which parameters to use, click on the wizard and the default parameters for the search type will be set.
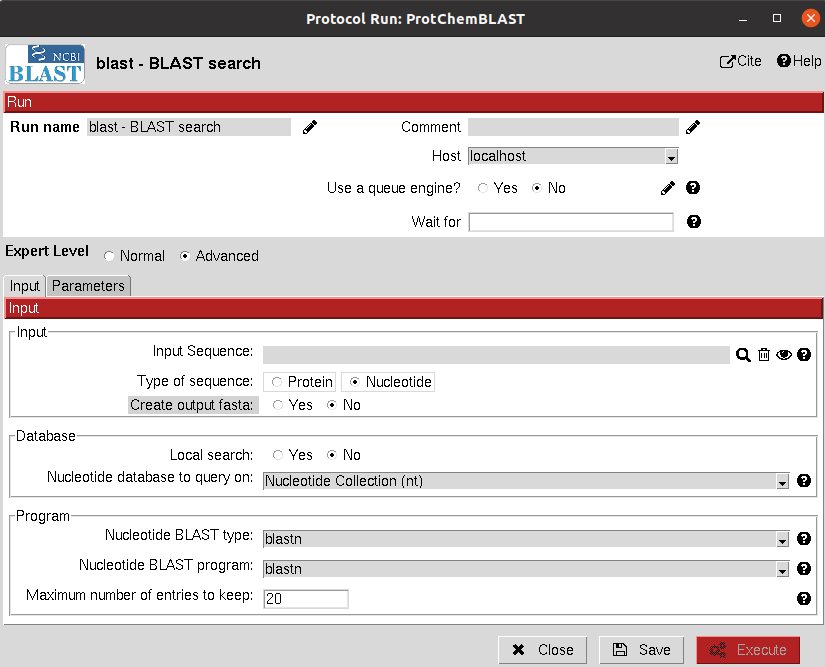
The result of this protocol is a SetOfSequences containing the BLAST hits. The analyze results button will open these sequences in AliViewer.
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests blast.tests.test_blast.TestBLAST