AutoDock-GPU Docking
This protocol uses AutoDock-GPU:, the GPU implementation of AutoDock, which is several times faster and includes many bug fixes and new features.
It includes different parameters than AutoDock4, so it has been moved to a new protocol.
The input can either be an AtomStruct (to perform the docking on the whole protein) or a SetOfStructROIs (to perform the docking only on the Structural Regions Of Interest).
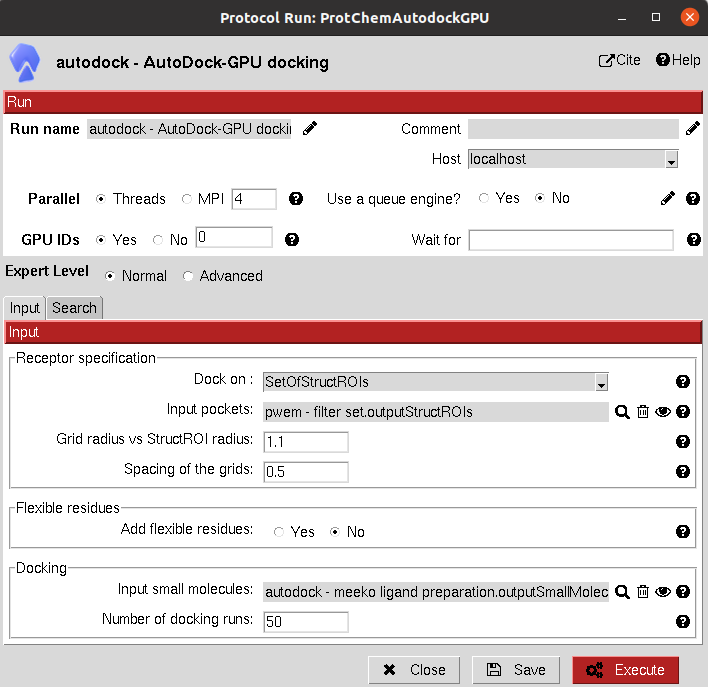
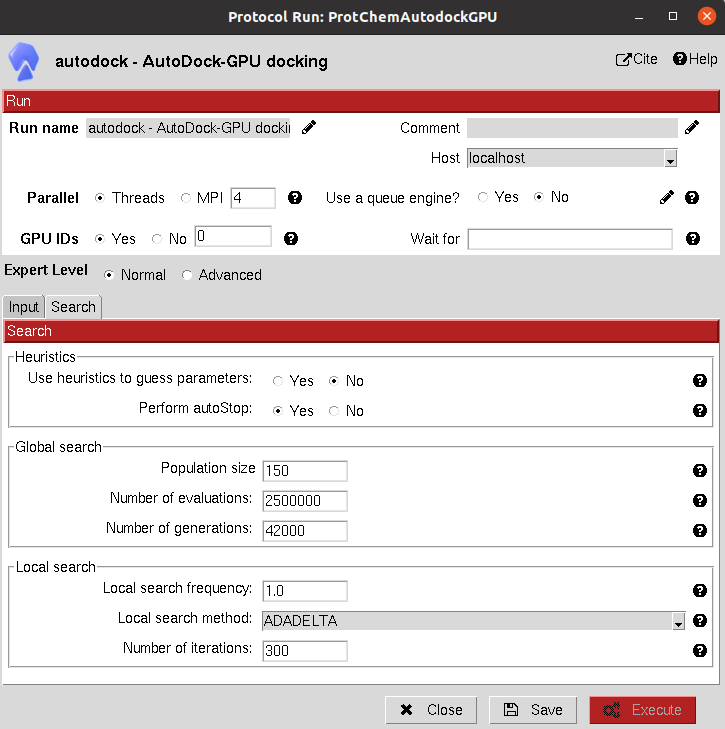
The results of these protocols are a SetOfSmallMolecules, containing the predicted binding poses for the input molecules.
The user can visualize them using Analyze Results, which will display the General SmallMolecules viewer.
A section for defining flexible receptor residues is included in these docking protocols.
Test
This protocol has an integrated test that can be run using the following command:
scipion3 tests autodock.tests.test_autodock.TestAutoDockGPU